Customer Success Story
How SMRT Sequencing Helped Researchers at University of Washington Uncover a Tandem Repeat Linked to ALS
PacBio long-read sequencing removes the barrier of sequencing long tandem repeats found to be linked to ALS.
- The ProblemMore than 40 tandem repeat expansions in the human genome are known to cause neurological disease. Short-read sequencing cannot capture these large repeat expansions, causing them to be missed in many studies.
- The SolutionHighly accurate long-read sequencing (HiFi sequencing) allows scientists to capture the full range of genomic variation - even in challenging regions of the genome like large repeat expansions.
- The ResultThrough HiFi sequencing, researchers pinpointed a 69-bp VNTR in the WDR7 gene that was found to be enriched in three independent cohorts in individuals with ALS.
ALS at a Glance
Currently, the causes of ALS are largely unknown. Could the missing heritability be explained by complex genomic variants that are not captured with genotyping or short-read sequencing methods?
*in the United States
Uncovering a Genetic Variant Linked to ALS
A serendipitous discovery led University of Washington scientists to a tandem repeat that appears to explain some cases of sporadic amyotrophic lateral sclerosis, or ALS. This adds to the growing number of neurological diseases associated with tandem repeats, and provides a promising new path for identifying people at risk of developing ALS.
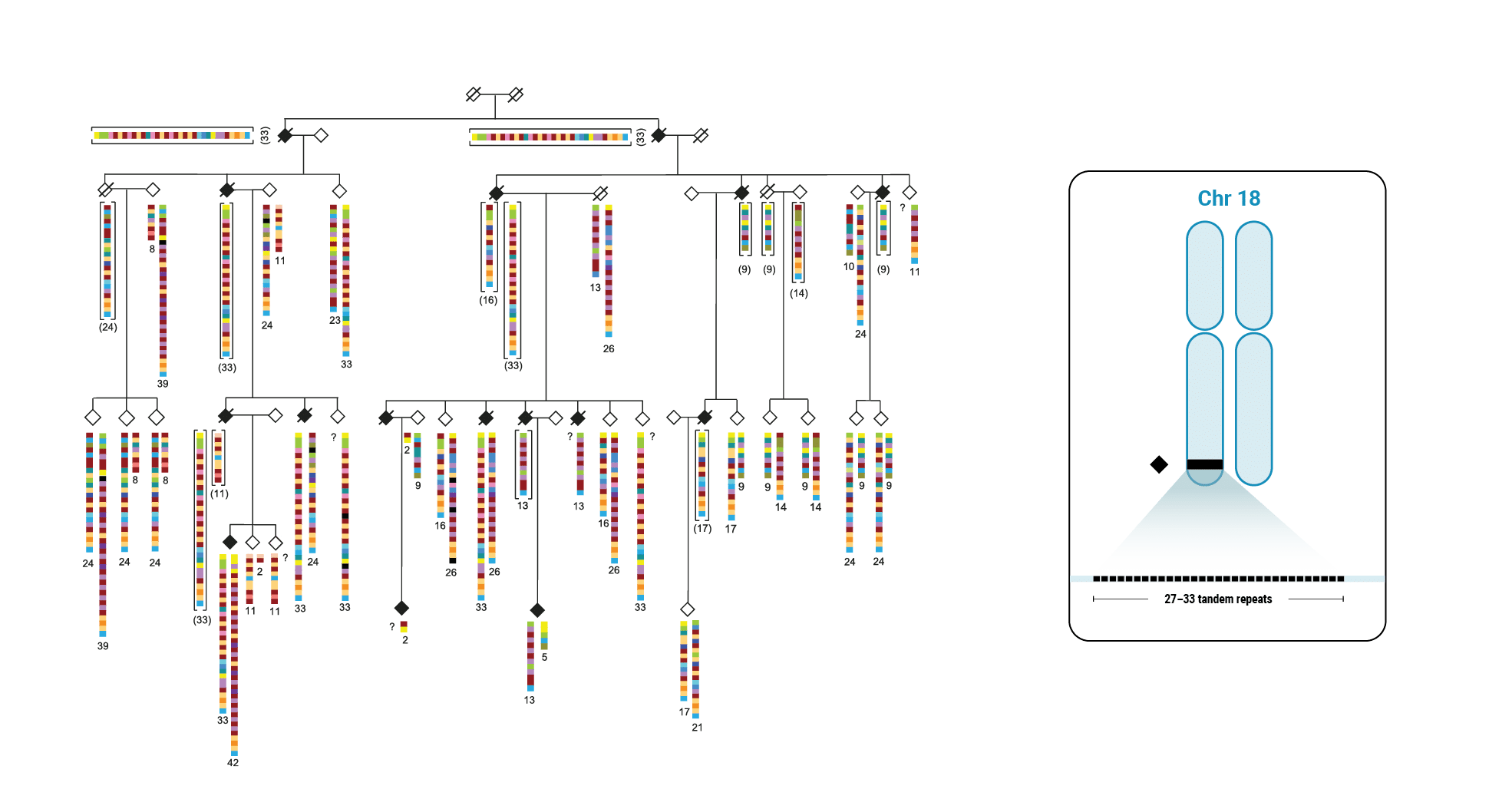
This discovery began with scientists interested in learning more about variable number tandem repeats (VNTRs). Paul Valdmanis (@pvaldmanis), assistant professor of medical genetics, and Meredith Course, a postdoctoral fellow in his lab, were pursuing the genetic cause of ALS in a large multigenerational family with several members who had developed the disease. That work pointed them toward a region on chromosome 18 that included a known tandem repeat, seen in a fairly expanded state in affected family members.
Multi-generational Family With ALS Affected Members

HiFi Long-Read Sequencing
VNTRs present a serious technical challenge for scientists. These elements can be extremely long, and with their repetitive nature, it is impossible to characterize them accurately using short-read sequencing technology and the necessary post-sequencing alignment process.
Benefits of Long-Read Sequencing
Highly accurate long reads, or HiFi reads, have allowed scientists to sequence through these challenging regions, base by base, for the most comprehensive and complete representation of even very large repeat expansions. Also known as Single Molecule, Real-Time (SMRT) Sequencing, this technology has shed light on repeat-associated neurological diseases such as fragile X syndrome, ataxias, and more. These discoveries clearly show that both the number of repeats and any tough-to-spot base interruptions in the region can be essential to understanding which people may develop a disease and which may not.
Course and Valdmanis chose to implement SMRT Sequencing for their ALS study and wound up revealing far more than they expected. “Once we decided to pursue long-read sequencing, we were really surprised to see the patterns that arose,” Course says.

Study Highlights
The scientists focused on a gene called WDR7, where a large, 69-base intronic repeat appeared to meet their criteria for a possible mechanism associated with spontaneous ALS. In the affected family, people with ALS had about 27-33 copies of the tandem repeat.
Using a multiplexed sequencing strategy with barcoded amplicons, the team pooled nearly 300 samples and analyzed the WDR7 gene in each.
The VNTR was not what they anticipated. “We call it a living, breathing repeat,” Valdmanis adds. “It was actually growing in one direction. As it’s building, it’s adding these new repeat units, two at a time.”
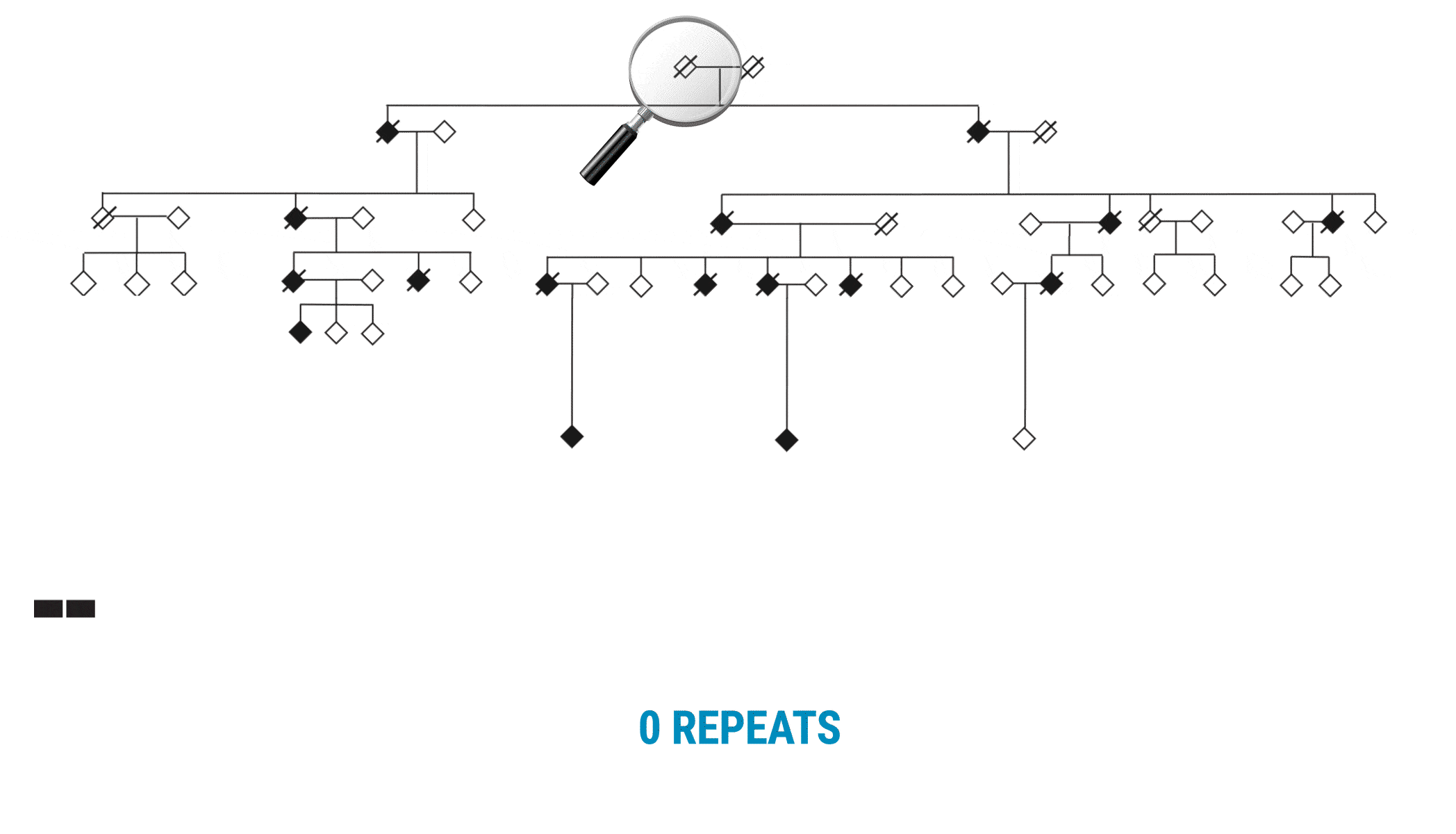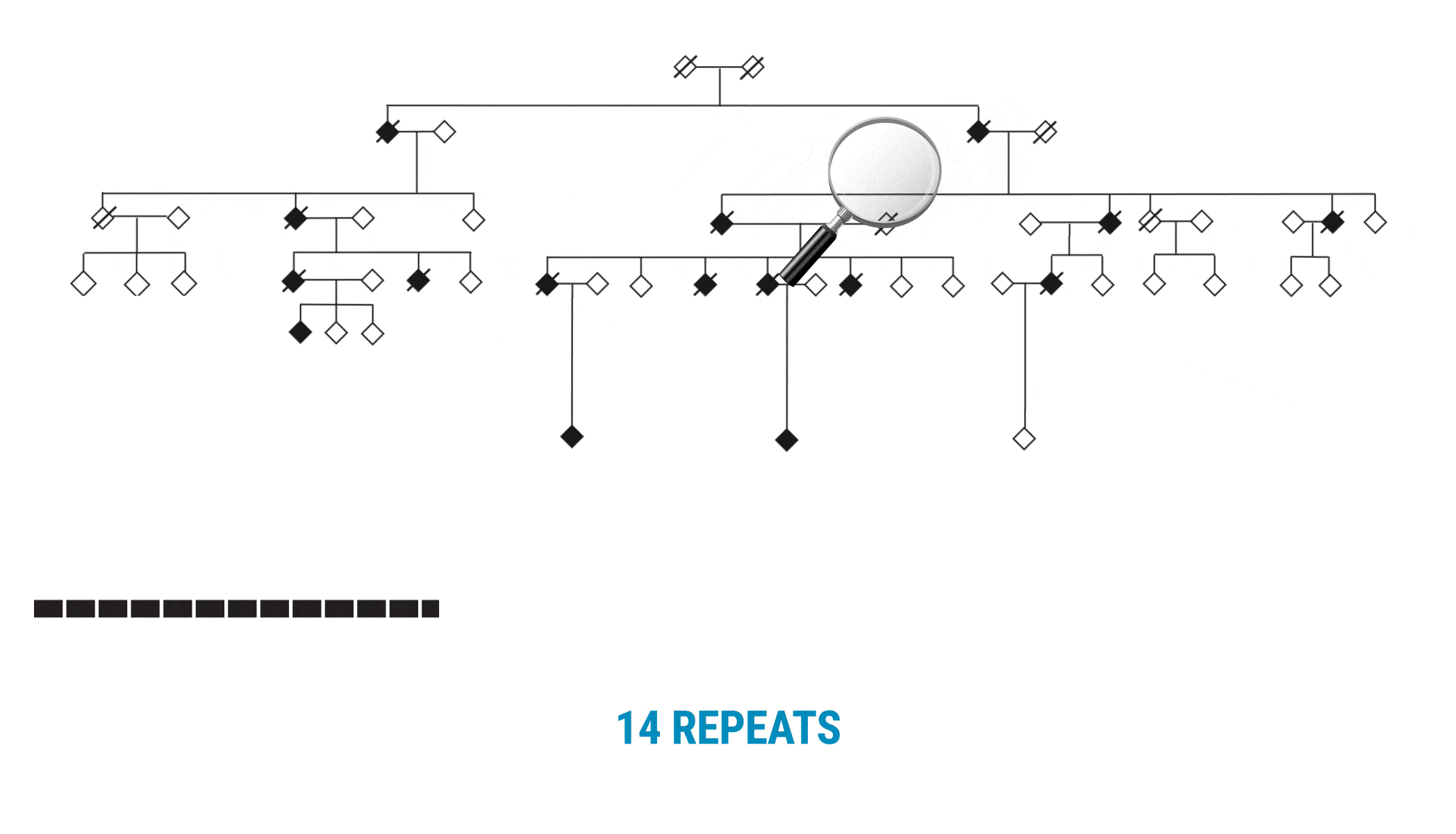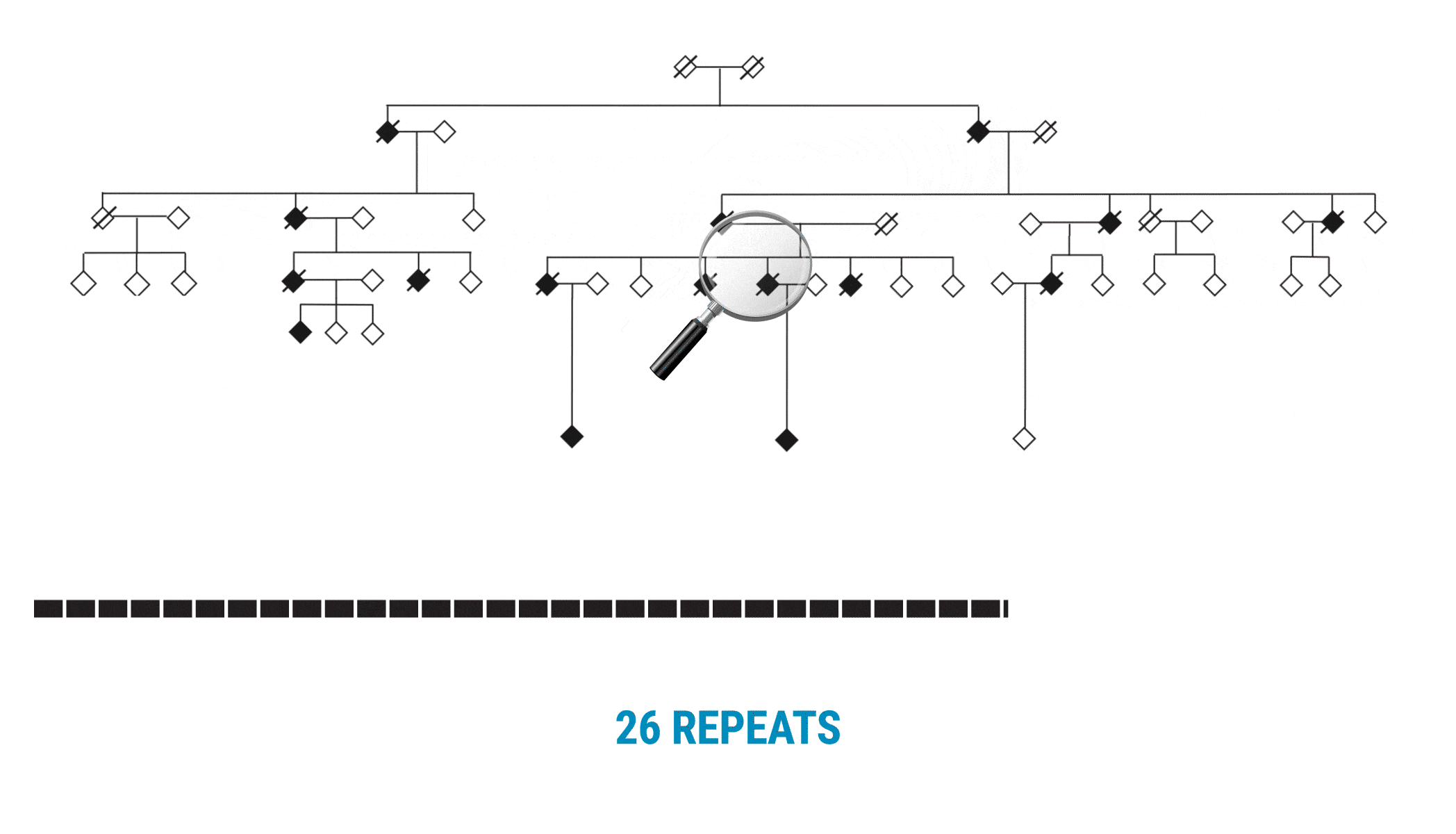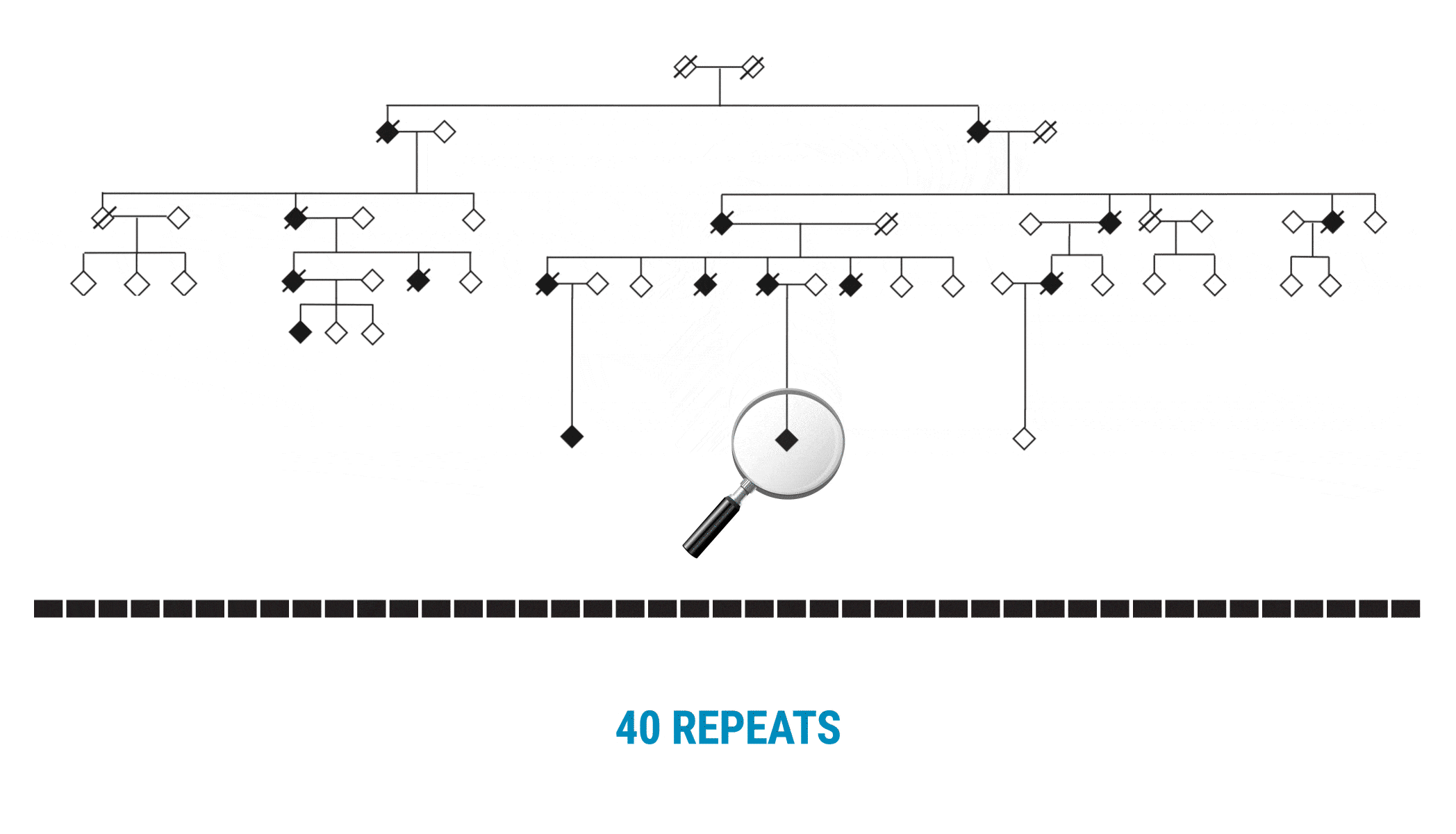

With SMRT Sequencing, the scientists had no trouble getting an accurate count of the repeat in each sample. Perhaps more importantly, they were able to identify six of the 69 bases that varied across the repeat expansion. Those bases, and only those bases, changed across the entire VNTR. What they discovered was a clear pattern of variability or ‘interruption sequences’. “That gave us confidence that we had a very accurate method to call these nucleotides across the repeat,” Valdmanis says.
“With most research in tandem repeats, what they’re measuring is the length of the repeat,” says Course. “What long-read sequencing gives us is the internal nucleotide structure. That’s just unprecedented, and it’s extremely valuable.”
An in-depth investigation found that each repeat unit forms a structure that has the potential to produce microRNAs, as the scientists report in their publication in the American Journal of Human Genetics.

The team could have stopped there, but instead pressed on to learn more about this intriguing tandem repeat. They used data from the 1,000 Genomes Project to chart patterns of the VNTR across geographic populations, finding that not all internal repeat motifs are present in all groups. In addition, they considered the evolutionary history of the VNTR. The sequence is present in no more than a single copy in our bonobo, chimpanzee, gorilla, and orangutan cousins, so the repeat expansion appears to be specific to the human lineage. However, it appears in expanded form in Neanderthal and Denisovan genomes, so it must have begun expanding before the emergence of modern humans.
Evolutionary History of VNTRs

What This Means for the Future
“This is a really neat example of what so often happens in science, starting in one place and ending up somewhere else,” Course says. “We started out looking at something that was involved in disease and were surprised to find ourselves moving into population genetics and evolution.”
Valdmanis and his team hope that this study encourages other scientists to explore their VNTRs of interest with PacBio long-read sequencing. For their part, they plan to continue on this path in addition to their ongoing focus on understanding the role of the WDR7 repeat in sporadic ALS. “Now that we have a pipeline established and know how to amplify and understand the individual variation in repeats, we’re taking a look at other repeats that seem to have extended in a similar manner after the primate split,” he says. “We can use this technology to evaluate how repeats expand across the genome.”
It promises to be a fruitful quest. After all, tandem repeats appear to have strong association with diseases, particularly neurological ones. Many human-specific VNTRs are involved in synaptic transmission. “If we look at human-specific tandem repeats, these are areas of the genome that evolved fairly recently” — such as the brain, Course notes.
“These are genes that potentially could have expanded as drivers of evolution,” Valdmanis adds. “Perhaps the brain has an increased propensity for somatic expansion.”
Featured Neuroscience Content
Scientists Use HiFi Sequencing to Discover Variants Linked to ALS
Every May is ALS Awareness Month, and we’re hoping to help by spotlighting two deserving publications from scientists at the University of Washington and at the Mayo Clinic.Read Post
Raising Awareness for Ataxia & Understanding the Disease Mechanism
There are several hereditary ataxias, and genetic testing is increasingly useful for pinpointing the exact type affecting a patient.Read Article
No-Amp Targeted Sequencing With PacBio SMRT Technology
PacBio No-Amp targeted sequencing uses the CRISPR-Cas9 system and enables scientists to access previously unsequencable regions of the genome.Learn More
咨询专家
如果您有疑问、需要查看订单状态或想要购买仪器,我们随时乐意提供帮助。
在本网页上注册,即表示您同意,并同意 PacBio 根据我们的隐私政策收集和使用该信息.